Animating eyetools Data
Matthew Ivory, Tom Beesley
Source:vignettes/articles/Animating-eyetools-data.Rmd
Animating-eyetools-data.RmdIn this article, we offer examples of how eyetools data can be used with the gganimate package. This article is code heavy as it is designed to demonstrate the capacity of gganimate in tandem with eyetools and for relatively straightforward application to one’s own data.
data <- combine_eyes(HCL)
data_118 <- data[data$pNum == 118,]
data_118 <- interpolate(data_118)
data_smooth <- smoother(data_118, span = .02)
data_fix <- fixation_dispersion(data_smooth)Plotting raw data with lag
Using the plot_seq() function we can extract the raw eye
gaze and then we can create an animation using
transition_time() and shadow_wake() to provide
the ‘lag’ enabling the visualisation of the movement over the trial
time.
animate1 <- plot_seq(data = data_smooth, trial_number = 1,
bg_image = "../data/HCL_sample_image.jpg") +
scale_colour_gradient(low = "red", high = "red") + #to keep a consistent colour
guides(colour="none") + #remove redundant legend when colour is consistent
transition_time(time) +
shadow_wake(.125, wrap = FALSE, size = 2, alpha = .75)
animate(animate1,
duration = 15,
end_pause = 15)
## To save an animation
#anim_save("figures/point_lag.gif", animate1, height = 810, width = 1440, duration = 12,
# end_pause = 15)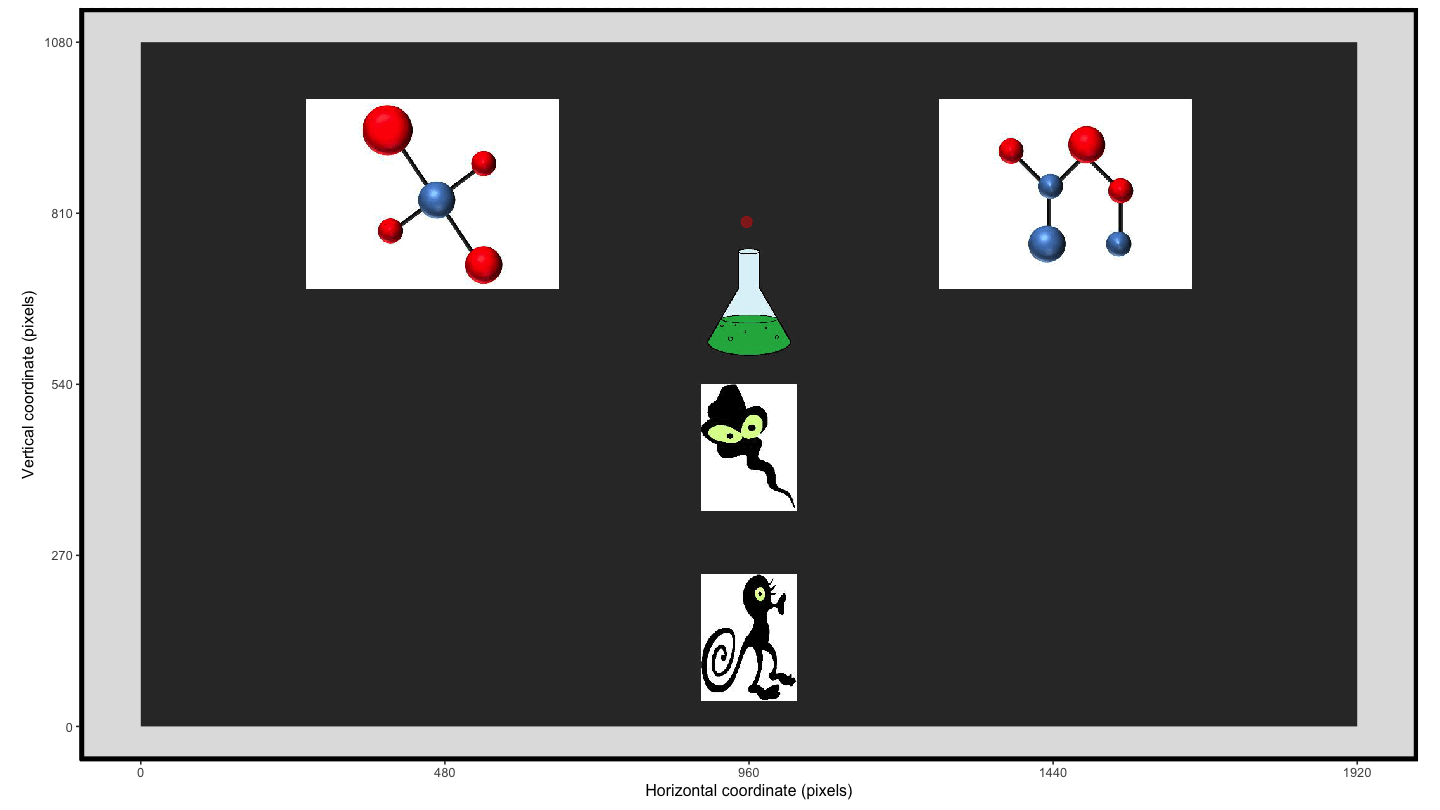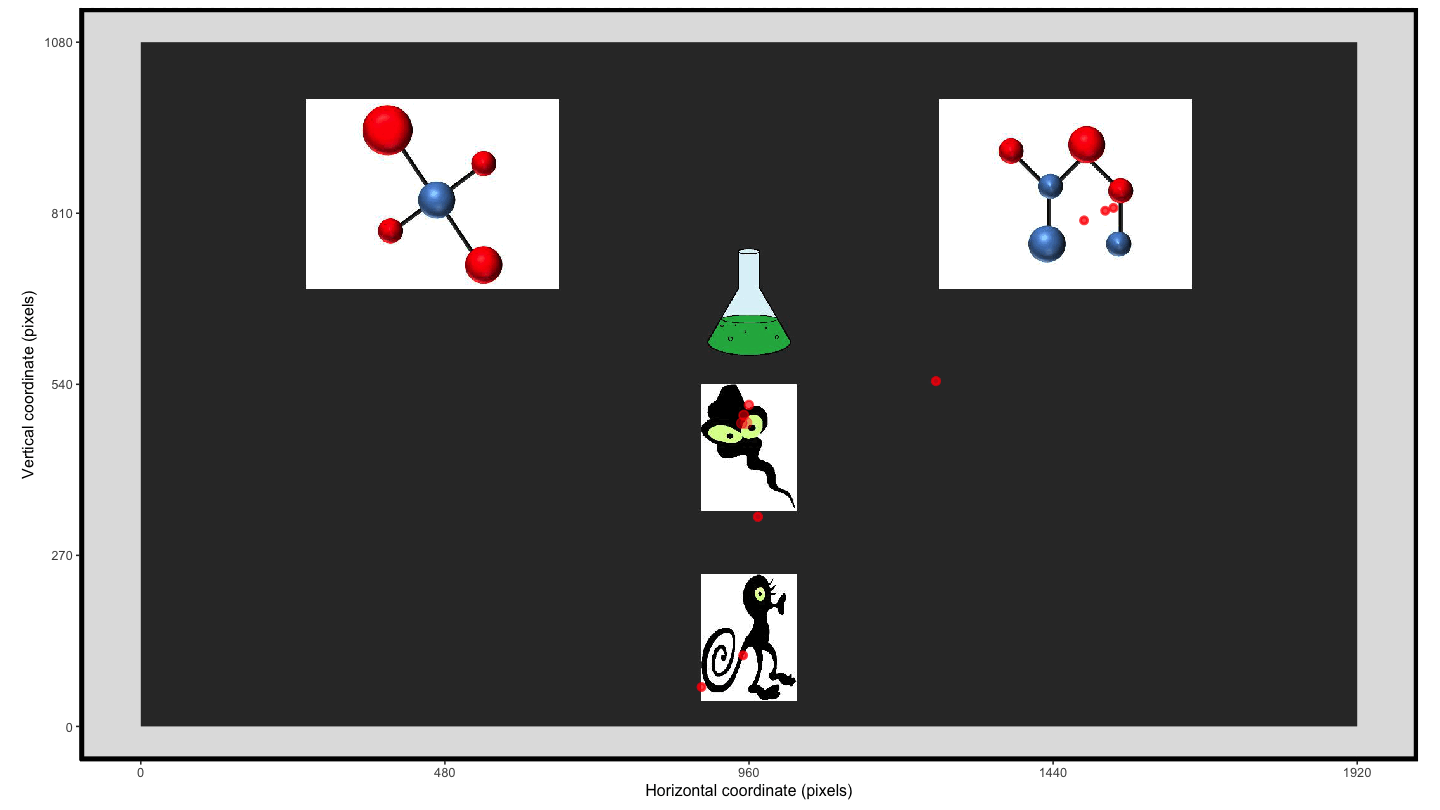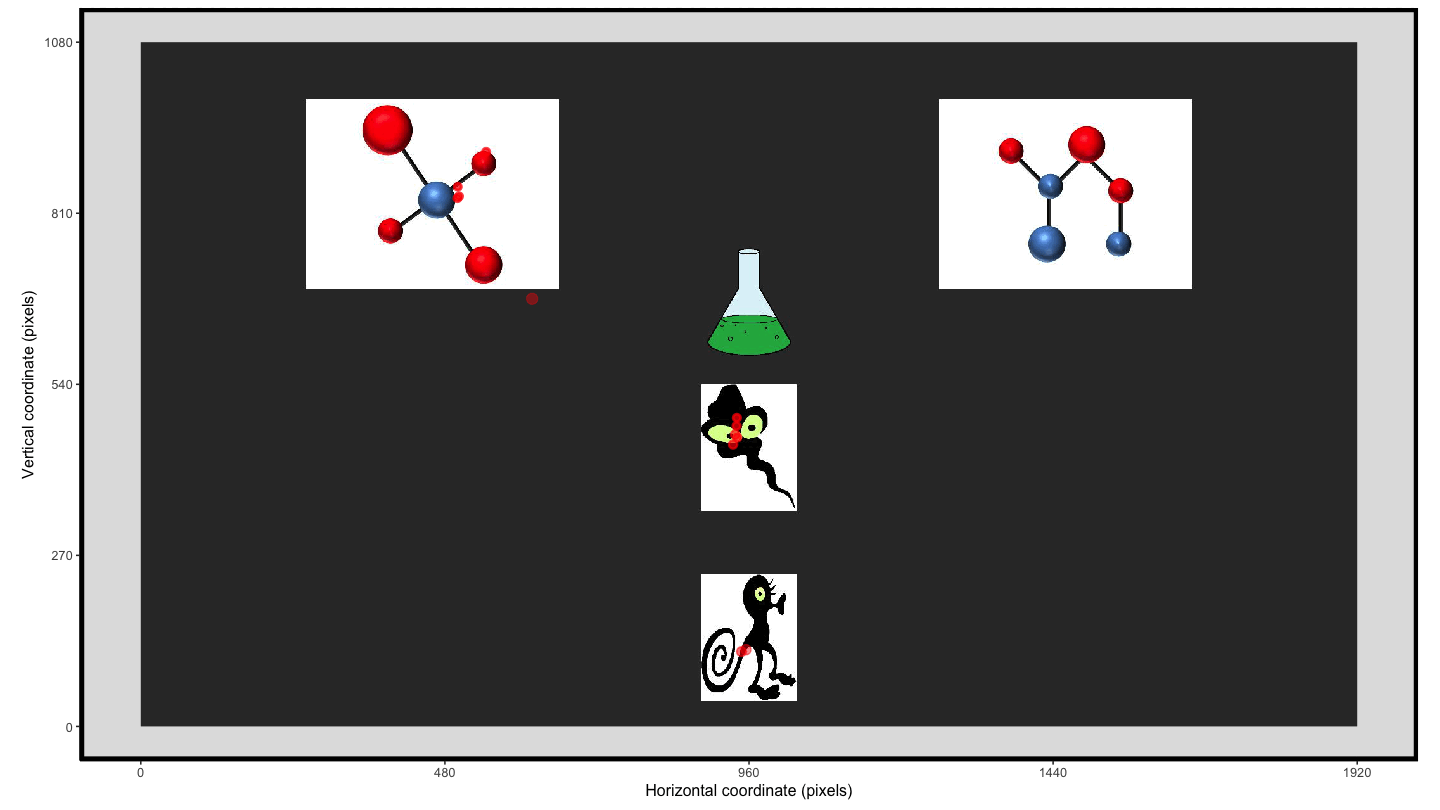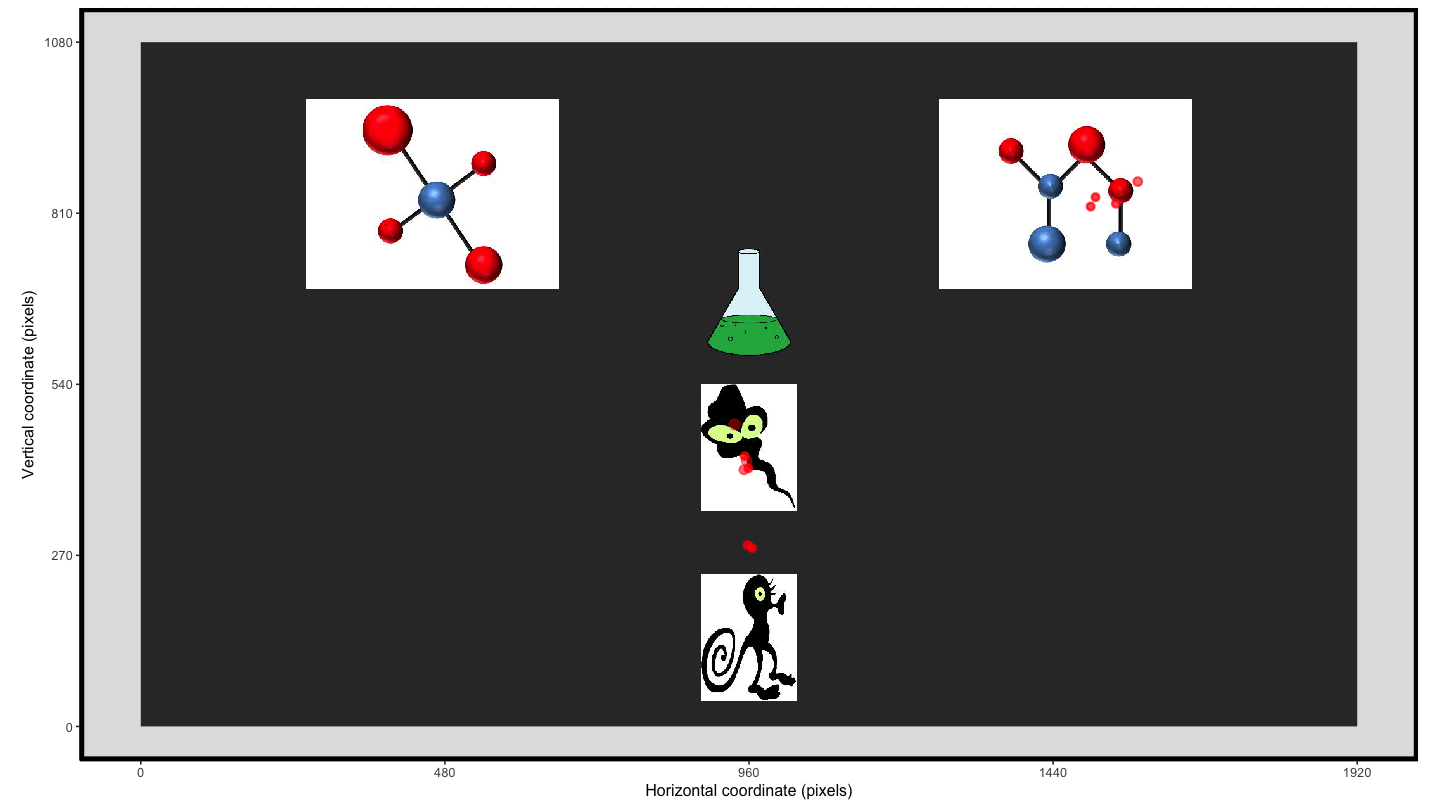
Plotting raw and smoothed data
We can also animate the static plot presented in the
smoother() function by plotting the x and y coordinates
together. To do so, we cannot leverage the plotting function in
smoother() as it does not play nicely with the
gganimate() package, so we use the smoothed and unsmoothed
data in a pipeline.
data_both <- data |>
left_join(data_smooth, suffix = c("_raw", "_smooth"), by = join_by("pNum", "time", "trial")) |>
filter(pNum == "118", trial == "1") # take a single participant and trial
data_both <- data_both |>
pivot_longer(cols = c("x_raw", "x_smooth"), values_to = "x", names_to = "x_names") |>
pivot_longer(cols = c("y_raw", "y_smooth"), values_to = "y", names_to = "y_names") |>
filter((x_names == "x_raw" & y_names == "y_raw") | (x_names == "x_smooth" & y_names == "y_smooth")) |>
mutate(smoothed = str_remove(x_names, "x_"), .after = trial) |>
select(-c(x_names, y_names))
plot_animate_smooth <- data_both |>
mutate(alpha = ifelse(smoothed == "smooth", .5, 1)) |>
ggplot(aes(x = x, y = y, colour = smoothed, fill = smoothed, alpha = alpha)) +
geom_point(size = 10) +
lims(x = c(0, 1920), y = c(0, 1080)) +
scale_alpha_identity() +
transition_components(time) +
shadow_wake(.25, wrap = FALSE, size = 2, alpha = .75)
animate(plot_animate_smooth,
duration = 15,
end_pause = 15)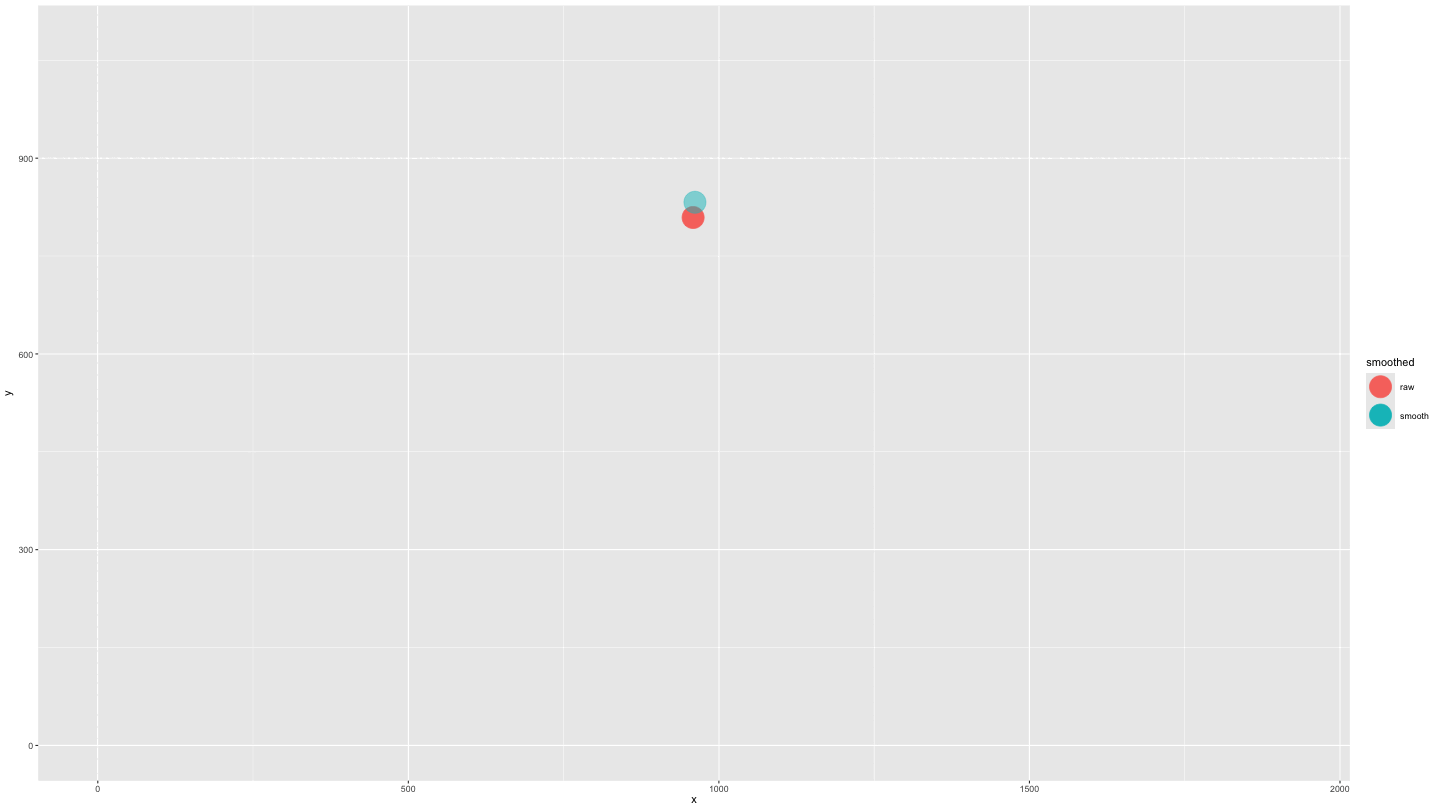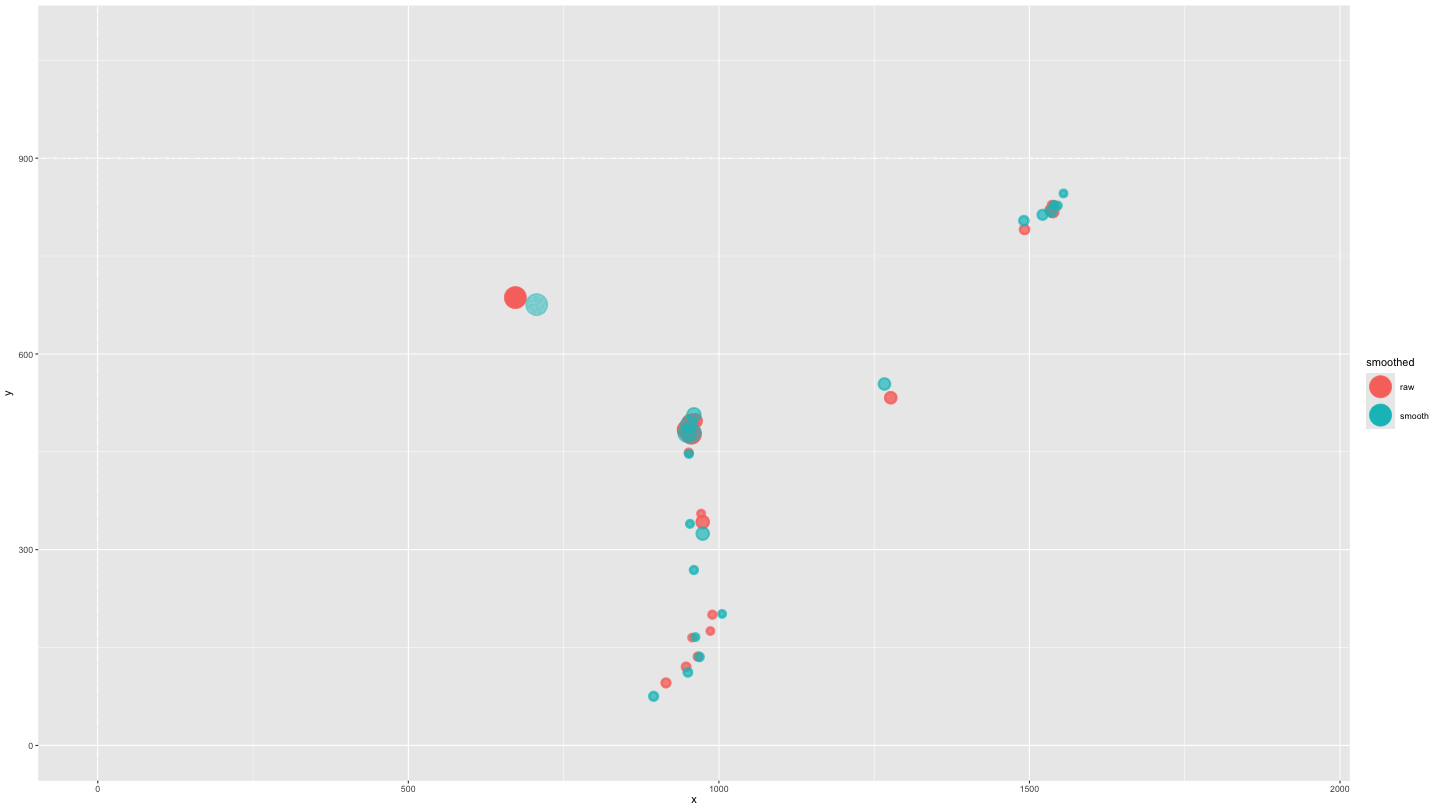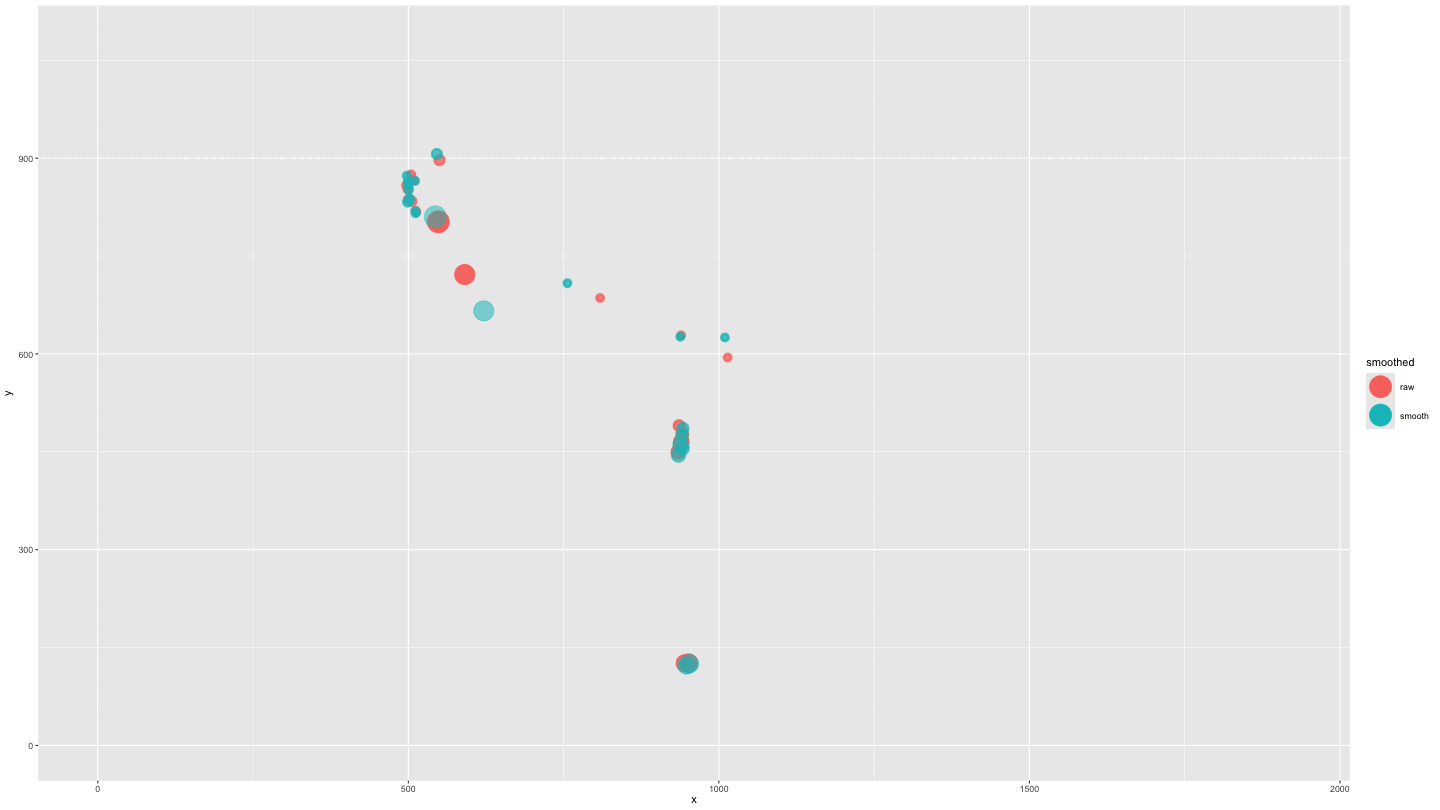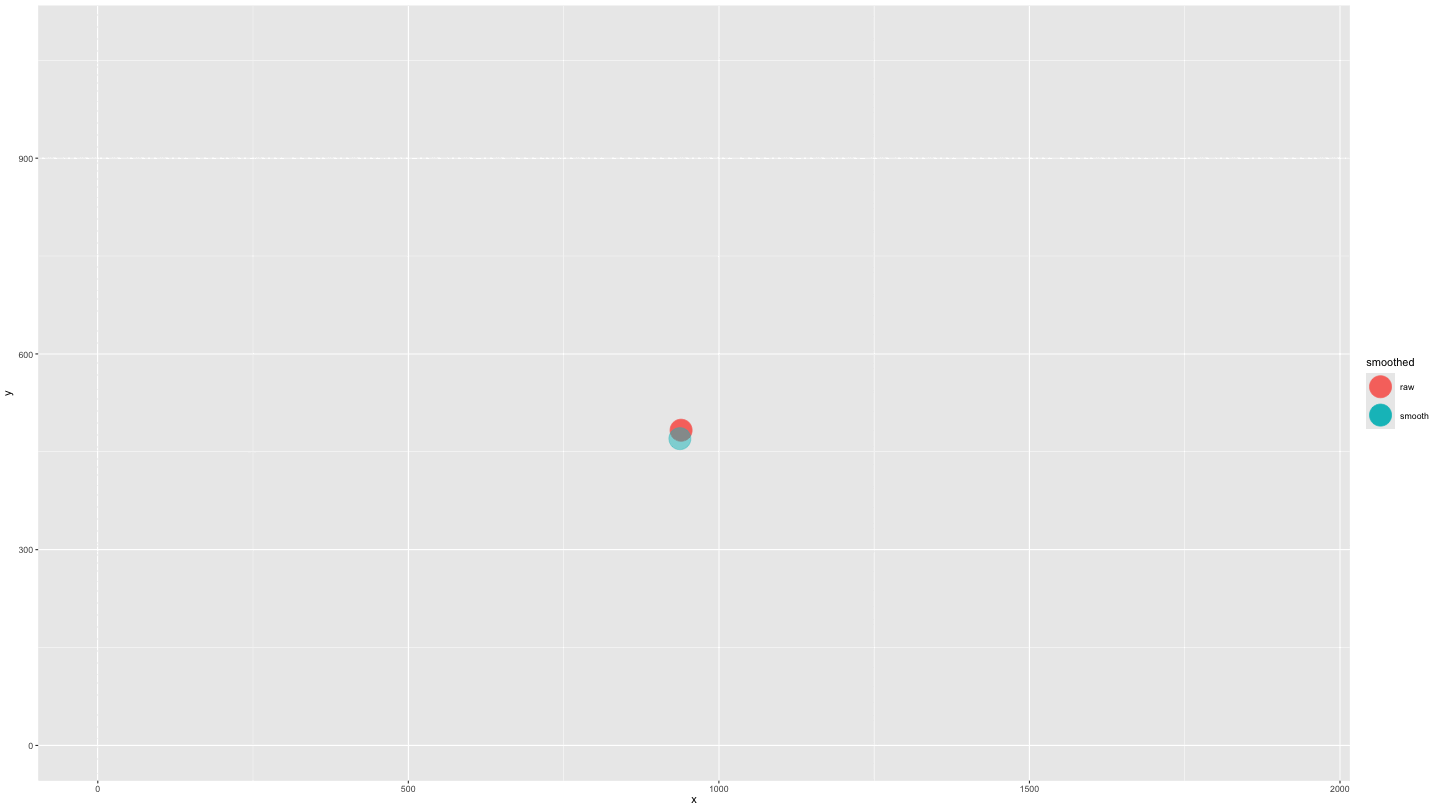
Plotting AOI entries
We can use AOI_seq() to get information about AOI
entries combined with the raw data to show the gaze data
entering/exiting AOIs.
data_plot4 <- AOI_seq(data_fix, AOIs = HCL_AOIs, AOI_names = c("Predictive", "Non-Predictive", "Target")) |>
filter(trial == 1) |>
select(-c(trial, duration)) |>
pivot_longer(start:end, values_to = "time") |>
full_join(data_smooth |>
filter(trial == 1)) |>
arrange(time) |>
fill(AOI, entry_n, name, .direction = "down") |>
mutate(AOI = ifelse(name == "end", "out of AOI", AOI),
AOI = ifelse(is.na(AOI), "out of AOI", AOI)) |>
mutate(AOI = factor(AOI, levels = c("Predictive", "Non-Predictive", "Target", "out of AOI")))
plot_animate_4 <- data_plot4 |>
ggplot(aes(x,y, colour = AOI, group = pNum)) +
lims(x = c(0, 1920), y = c(0, 1080)) +
#add a background image
annotation_raster(magick::image_read("../data/HCL_sample_image.jpg"),
xmin = 0,
xmax = 1920,
ymin = 0,
ymax = 1080) +
geom_point(size = 5) +
transition_components(time)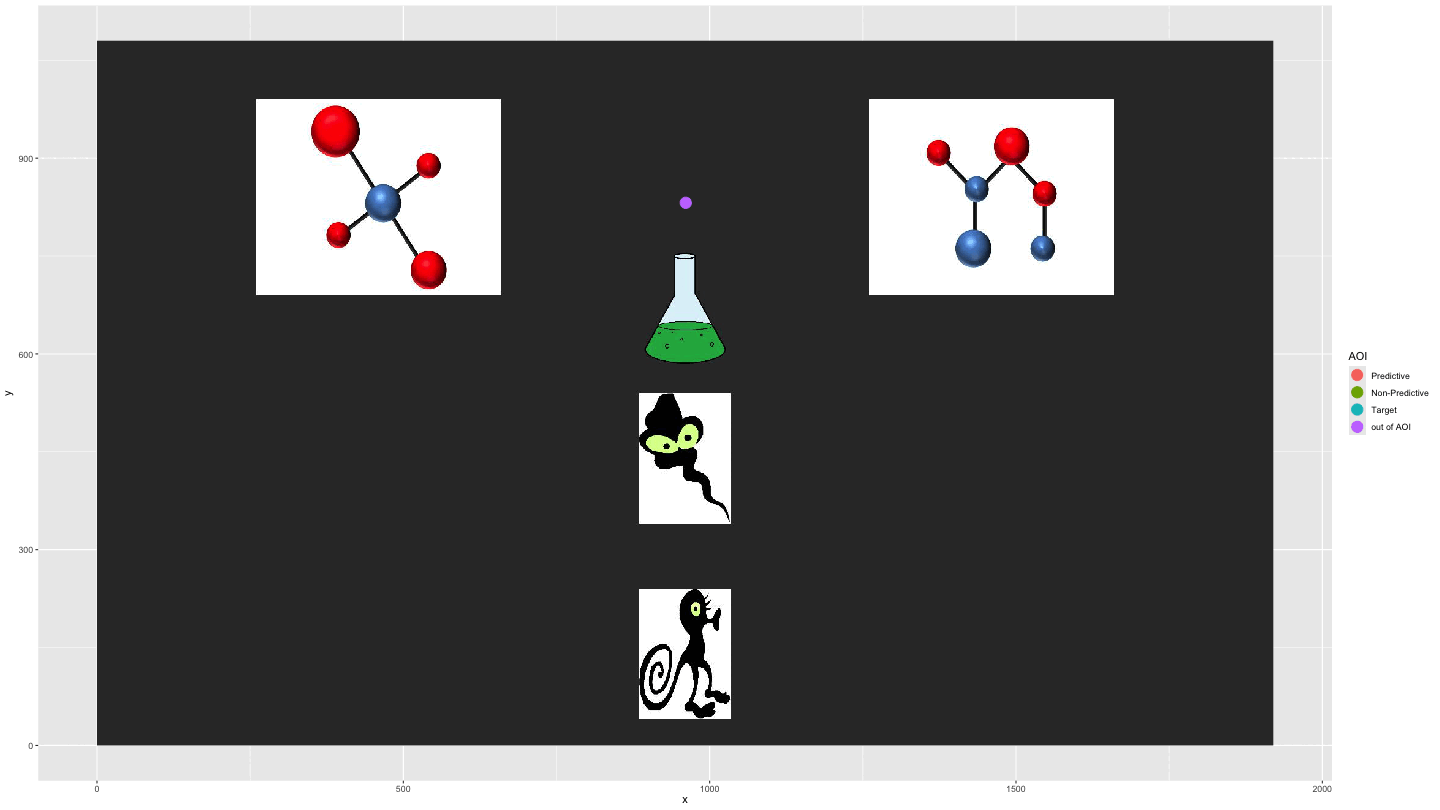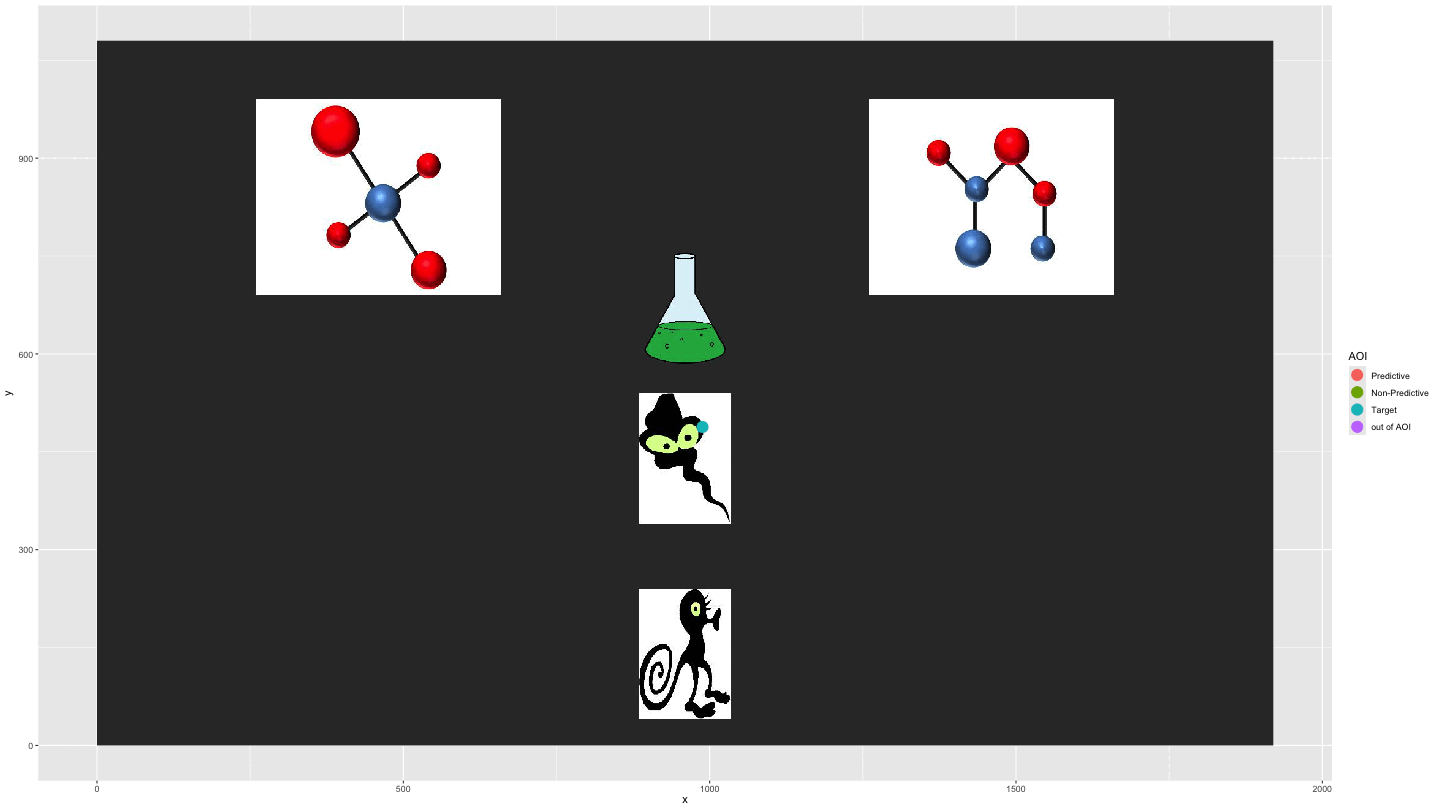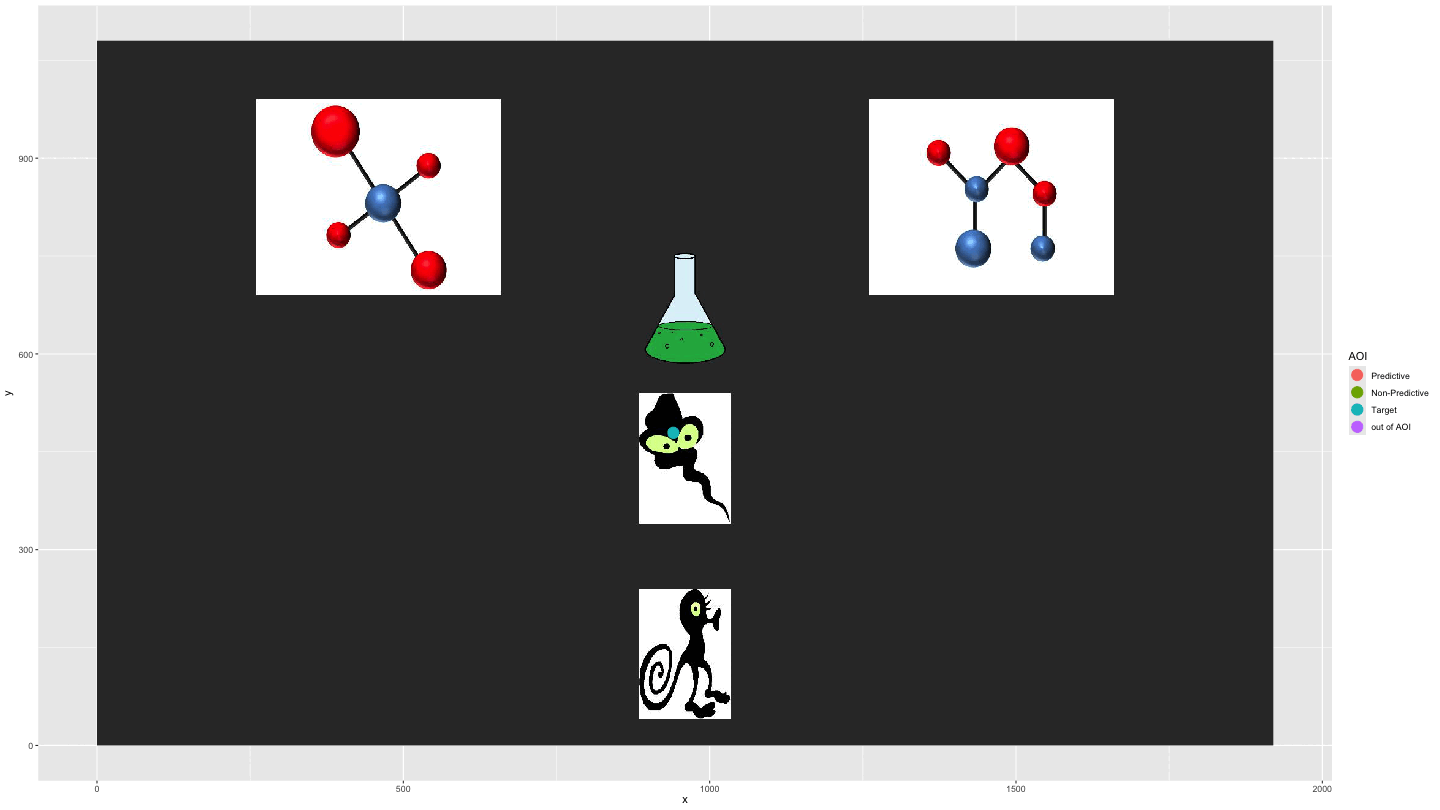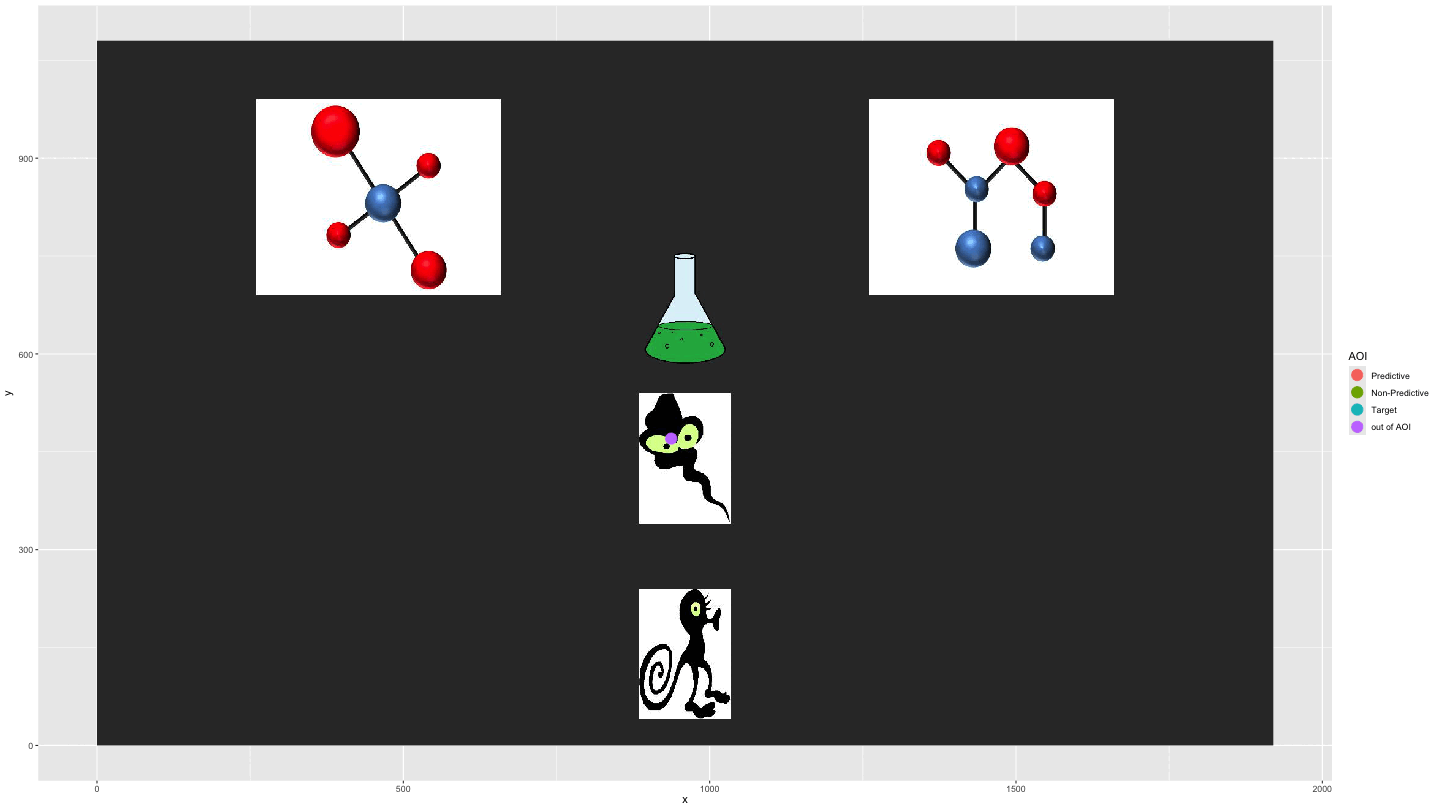
Plotting time spent in Areas of Interest over time
growth_all <- data_smooth |>
filter(pNum == "118", trial == "1") |>
plot_AOI_growth(AOIs = HCL_AOIs, AOI_names = c("Predictive", "Non Predictive", "Target")) +
geom_point() +
transition_reveal(time)
animate(growth_all,
duration = 10)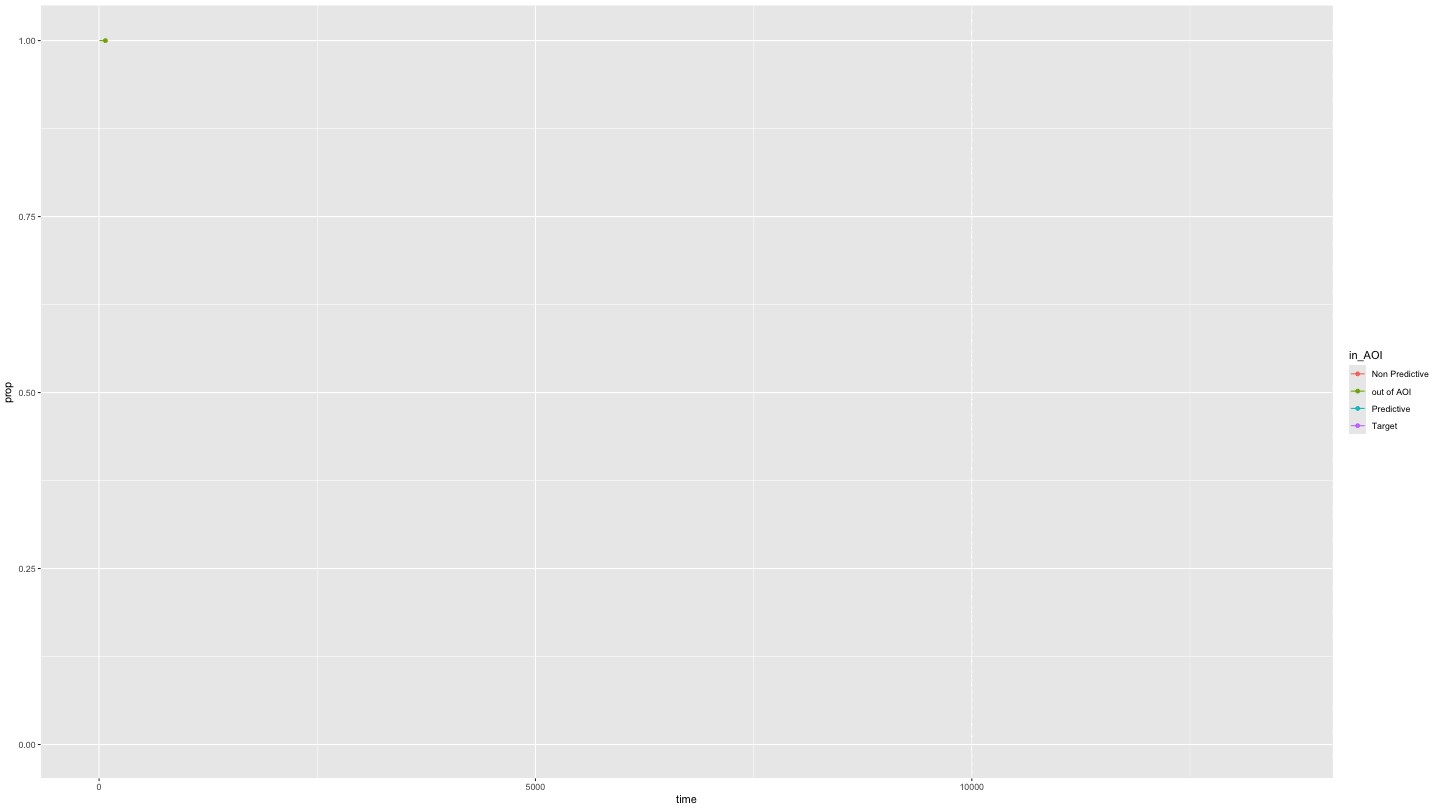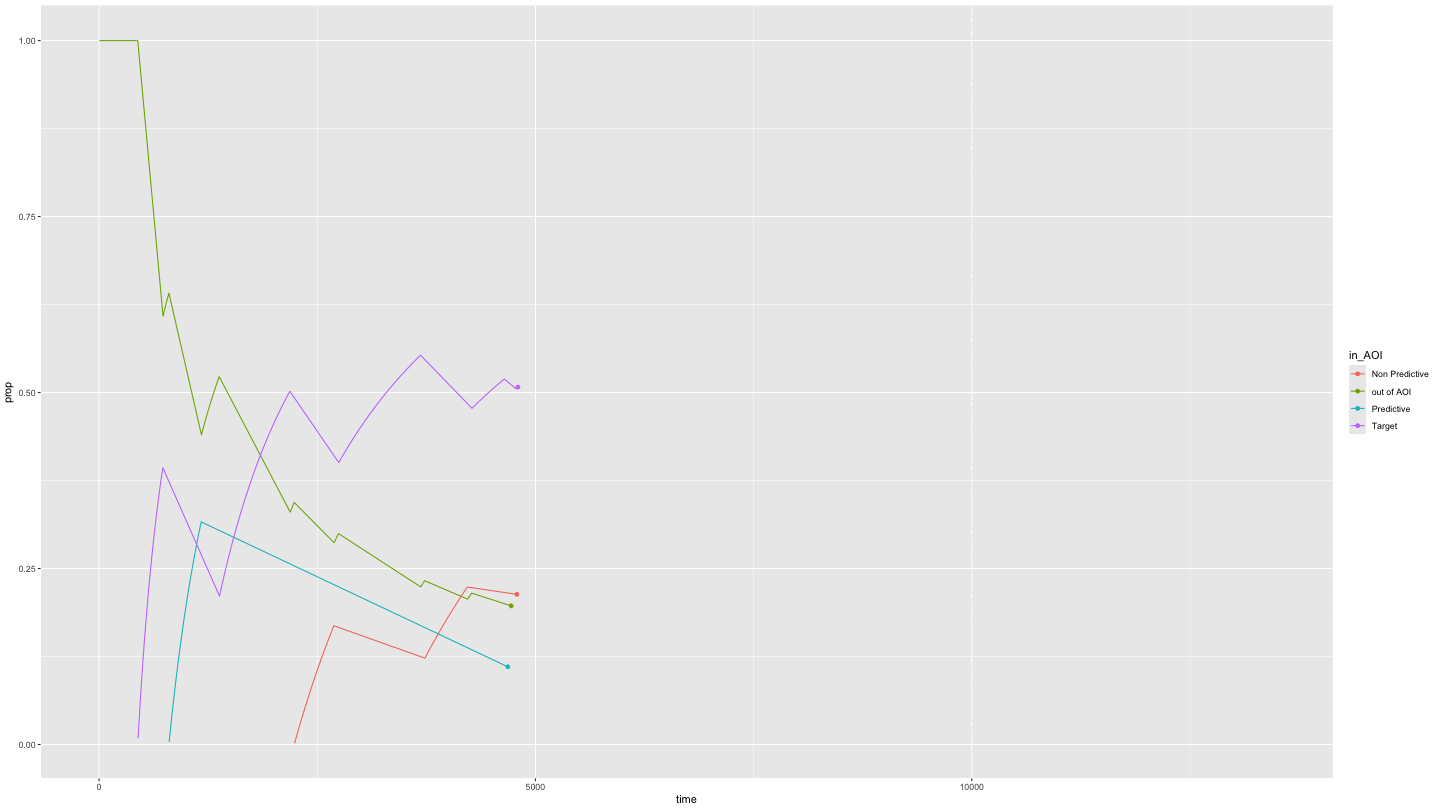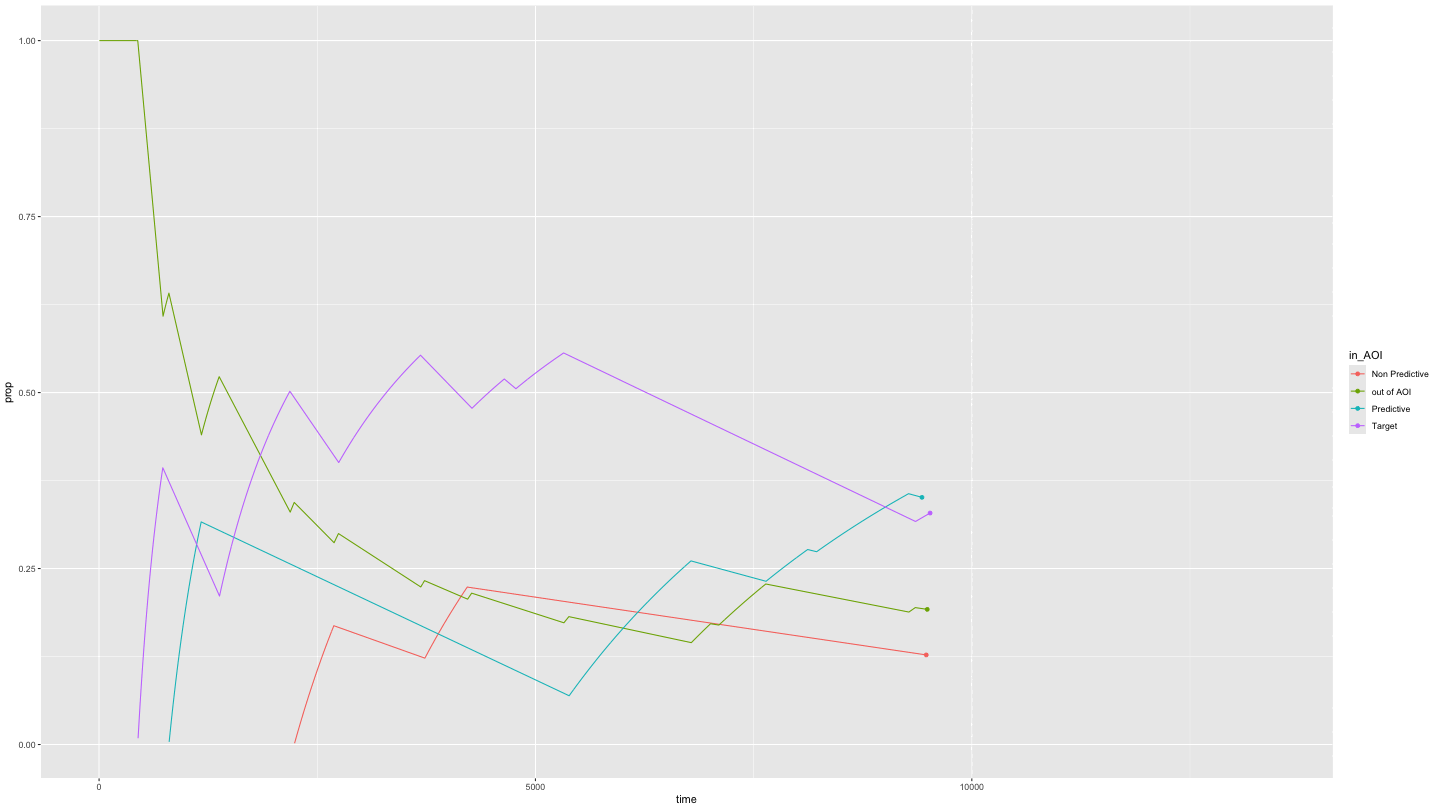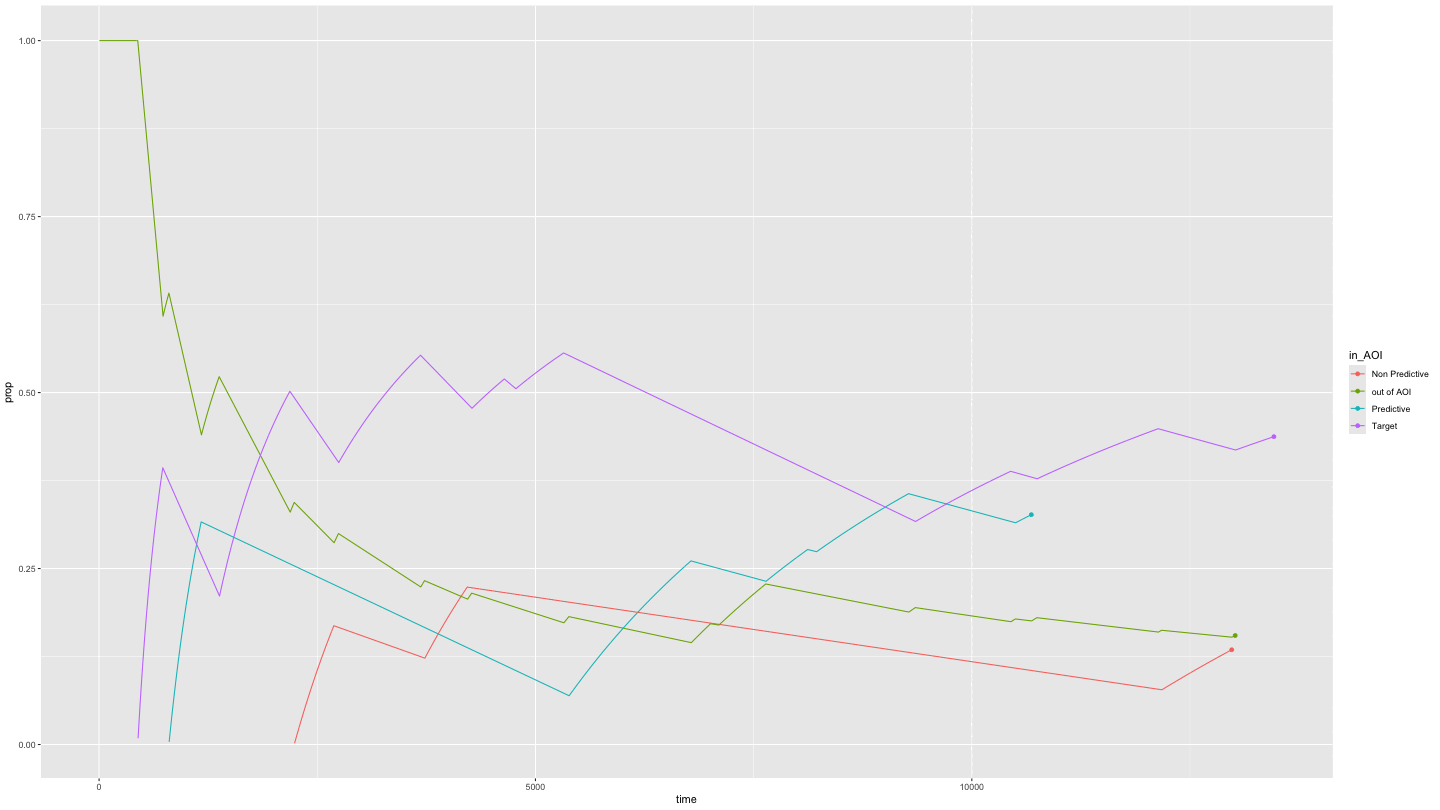
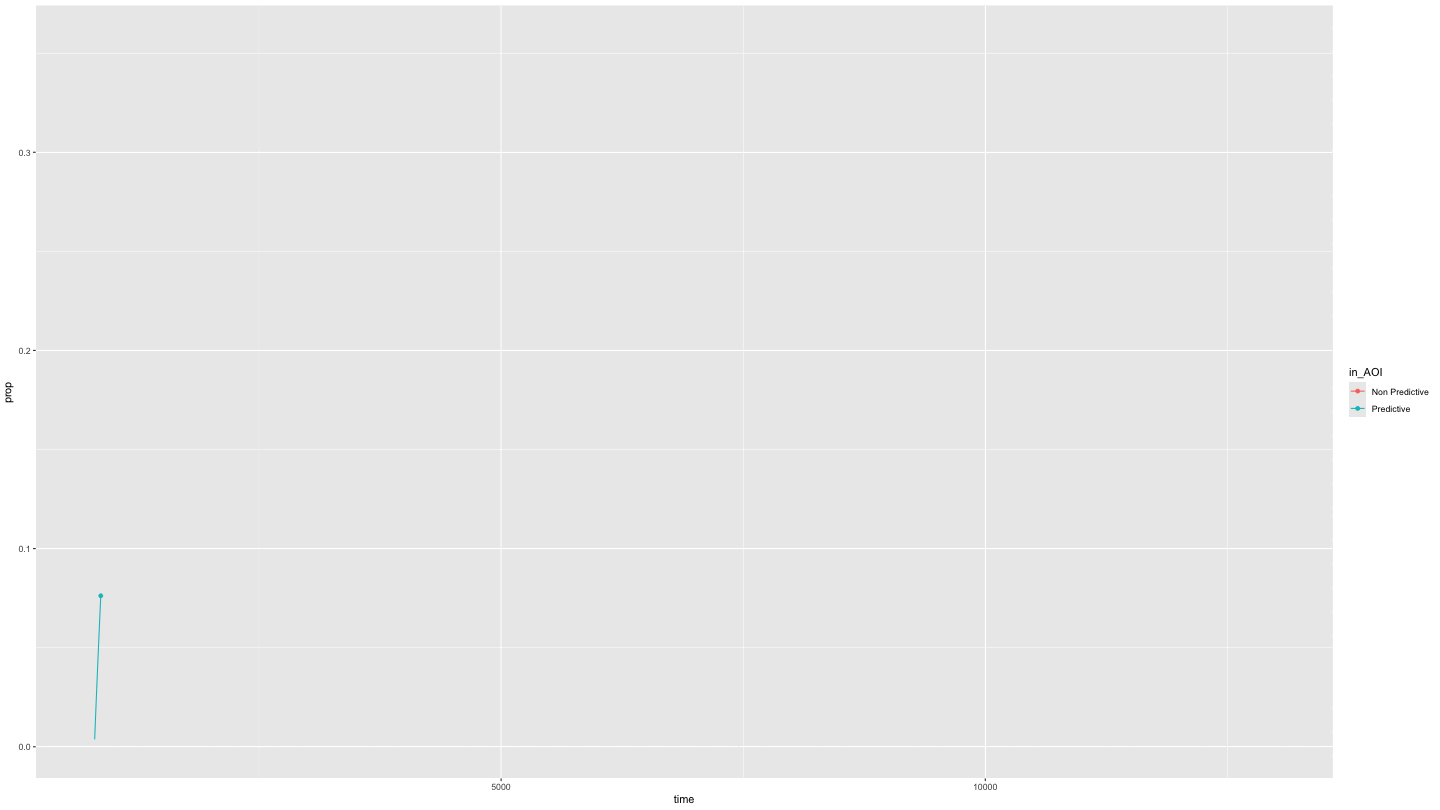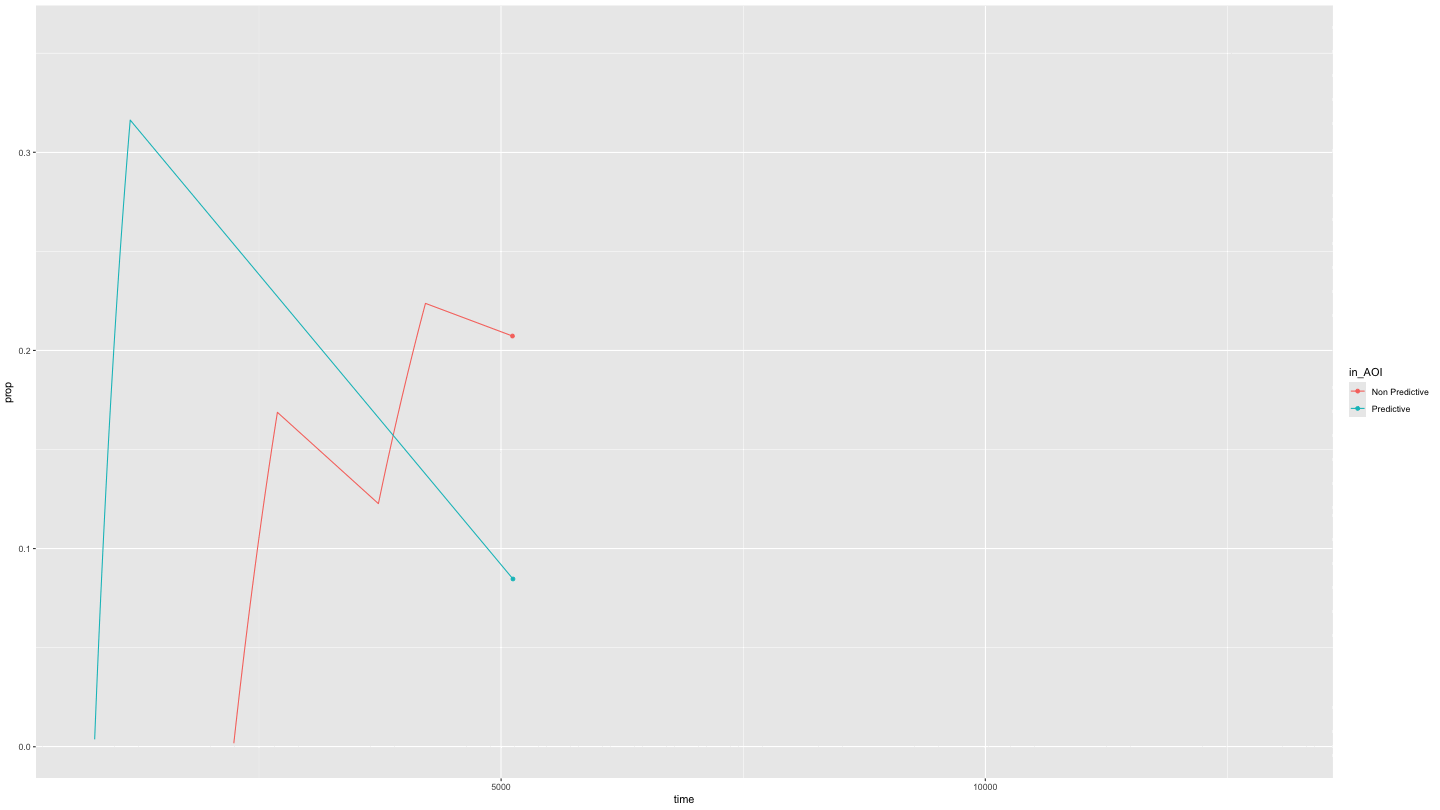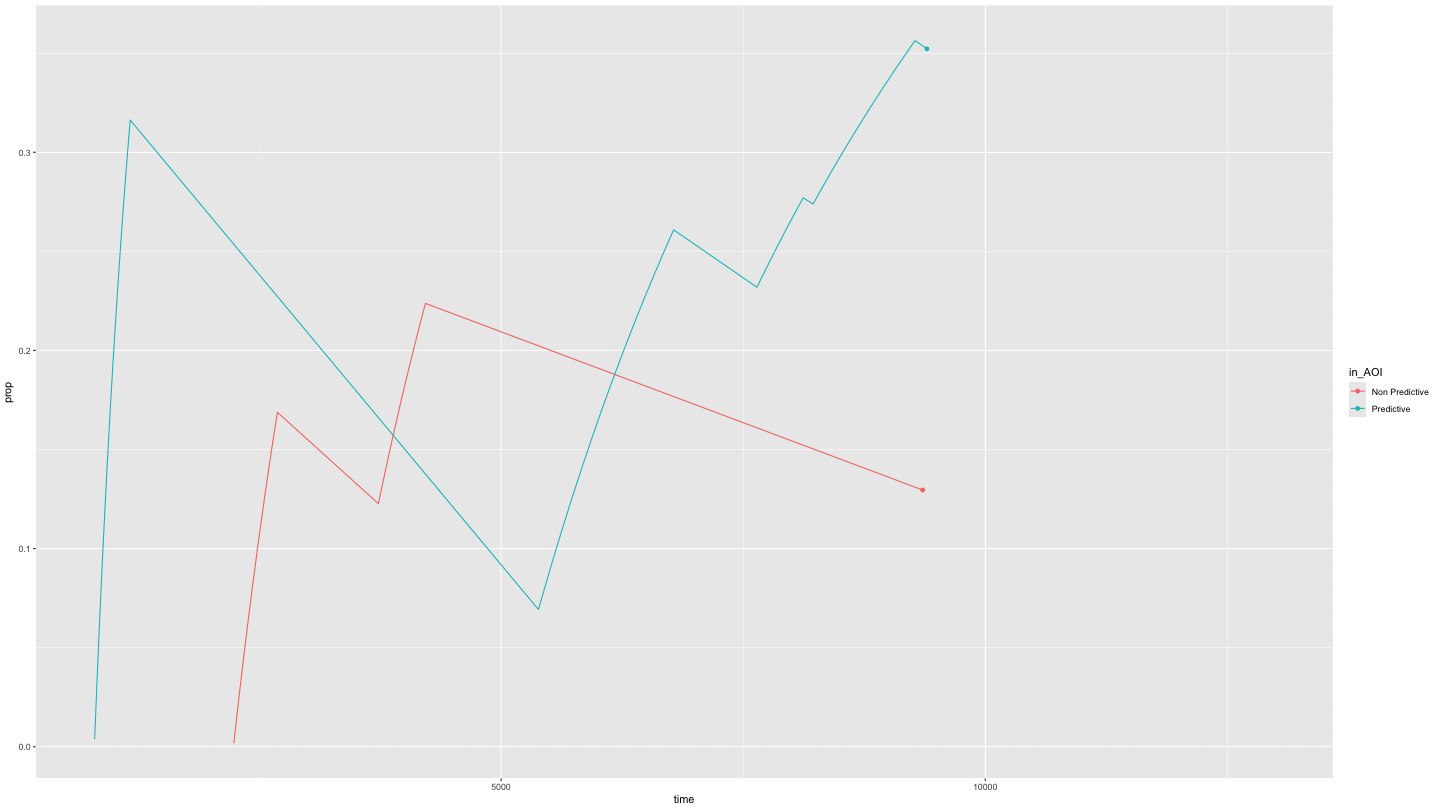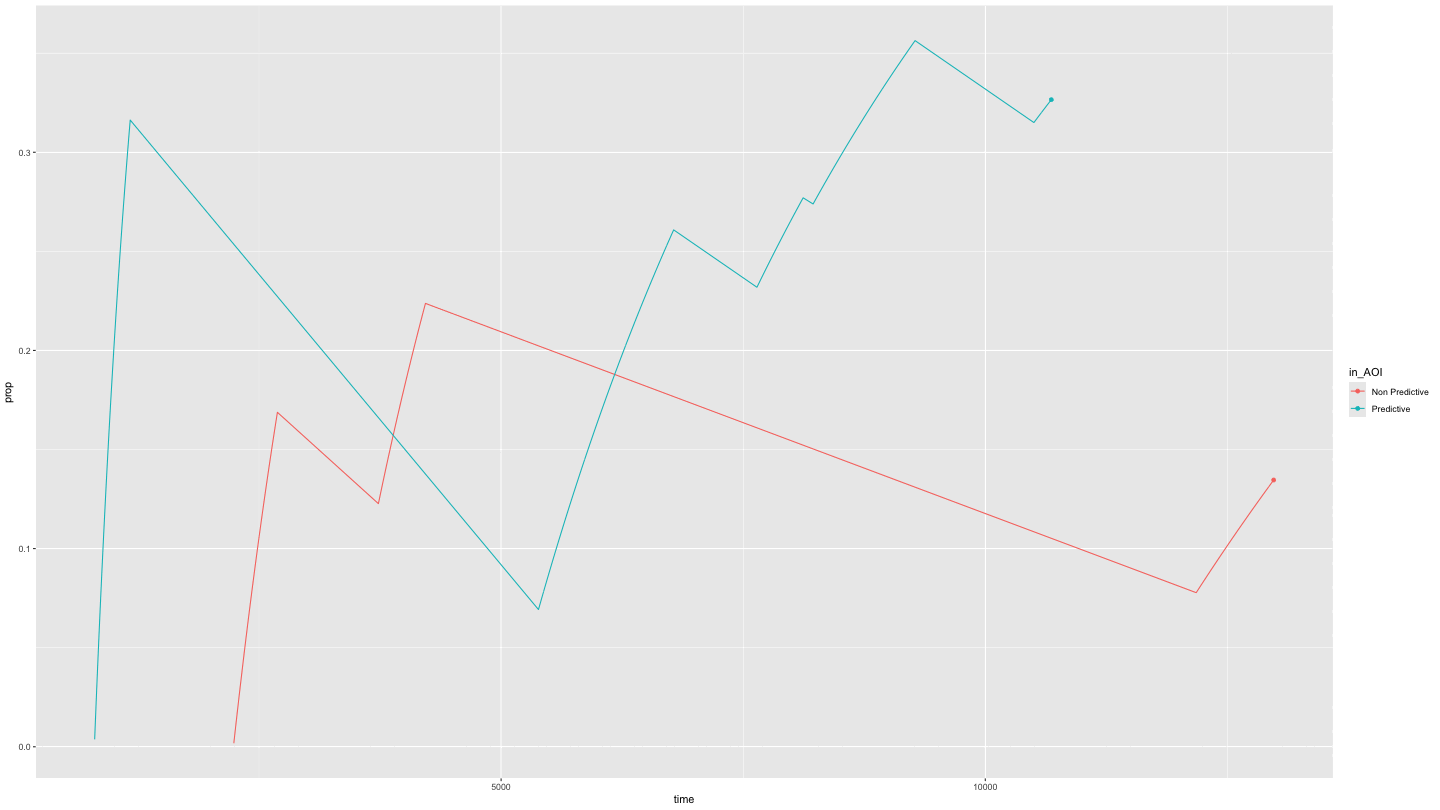
Or just predictive and non-predictive cues
This is easily done by specifying NAs in the appropriate position in
the AOI_names parameter.
growth_partial <- data_smooth |>
filter(pNum == "118", trial == "1") |>
plot_AOI_growth(AOIs = HCL_AOIs, type = "prop", AOI_names = c("Predictive", "Non Predictive", NA)) +
geom_point() +
transition_reveal(time)
animate(growth_partial,
duration = 10)